BEye Project
BEye was a project developed during my last year of bachelor at Politecnico di Milano, together with Lara Cavinato and Irene Fidone. It consists in an FPGA-accelerated application for retinal vessels segmentation, which aims at improving the performance (speed) of screening tests.
Context
Diabetic Retinopathy (DR) is an eye disorder due to diabetes compensation, and consists in the rapid growth of new anomalous vessels in the retina micro-circulation and the creation of micro-aneurysm and exudates or hemorrhages. It is a degenerative diseases that pass through three stages:
Non-proliferative with symptoms (such as edemas, micro aneurysms and a mild loss of sight/blurring) which can be detected by using screening tests
Proliferative stage, characterized by abnormal new blood vessels (neovasculari- sation) and bursted/bleeding and blurred vision with the presence of vitreous hemorrhages. In this phase, the situation gets more complicated and even the cures become more intrusive.
Blindness
The percentage of people who suffer from this pathology is more and more increasing, and screening tests are the best way to detect the disease before the proliferative stage. However, the algorithms used to identify DR are either too slow or costly to be extensively used. Here comes the opportunity tackled by BEye.
The project aims at making screening tests easy and fast, by using an embedded FPGA board with a fast vessels segmentation algorithm synthesized on it. Even if the work done by us was just a prototype of the algorithm synthesized on FPGA, it represents a good use case for FPGAs, and makes us think about its applications. For example, the FPGA could be associated with a high-speed camera to obtain a live segmentation and to check it with the previous images (or other similar patients). A mobile device like this would make screening tests easy to perform even where there aren’t near ophtalmologists and vision centers.
Vessels Segmentation Algorithm
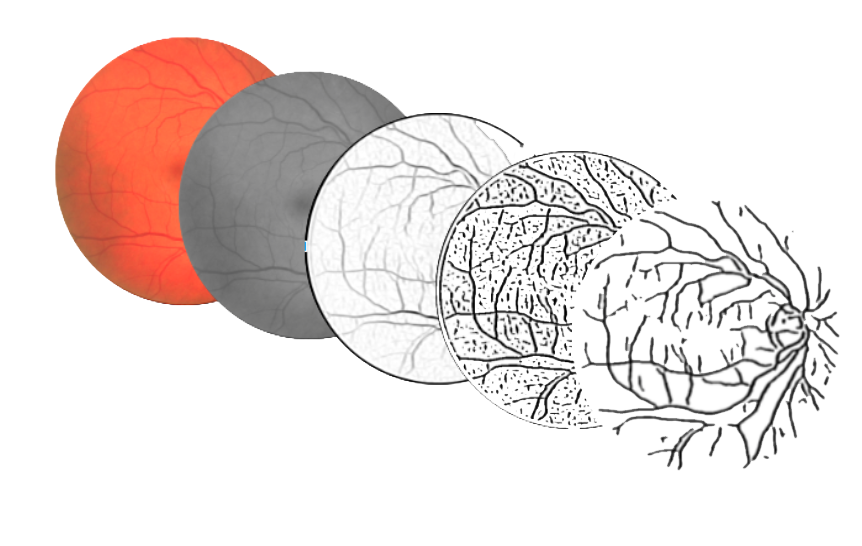
The algorithm for vessels segmentation performs a series of steps, each one implemented by a 2D filter on the input image.
The first phase is the preprocessing of the image with a 3x3 median blur, in order to remove salt and pepper noise which could impact on the next edge detection filters.
The vessel detection filter is instead implemented by performing a parallel filter of the image with 13 different kernels (16x16 sliding windows), taking the maximum value and then giving the output to a 9x9 adaptive threshold filter.
False positives are removed by employing a simple blob detection filter, implemented as a two-pass 2D filter over the image (takes the down and right pixels and get the maximum size connected component of black pixels).
Finally, the results are filtered for a last time with a 11x11 adaptive threshold.
Why FPGA?
As explained in the preceding description, the algorithm is composed of a series of 2D-filtering steps. A standard CPU (a single core) executes and runs one instruction at a time, and can perform one access to memory on each cycle. This means that, for each output pixel, the CPU would have to access the central and neighboring pixels each time, and even with a cache, it will mean K x K instructins for a single output pixel for a single filter, for each image.
An FPGA is instead like a blank canvas, in which instructions and local data accesses can be freely arranged (that’s why it this kind of execution is called spatial computation). The main advantage of using a FPGA in this project was then that it allows to arrange the on-chip fast memories (BRAMs) in order to parallelize the reading and execution of most of the filter operations. In this way, data locality is exploited by using the local memory for the local elements (neighbors), whithout waiting for the external memory (DRAM) each time.
The filters’ implementation takes advantage of this by creating a hardware sliding window (multipliers interleaved by registers and FIFO queues) in order to have a linear memory access (each pixel is accessed just one time from the main memory) for all the filters that can fit on the chip.
Also, the FPGA reconfigurability means that the design can be changed when the algorithm is updated, which happens often for biomedical applications.
Results
We created a prototype of the system by implementing the filters described in the algorithm section. We first implemented them in software (using python and the opencv libraries), then synthesized all of them for FPGA using Vivado HLS and Vivado IPI.
Vivado HLS is a high-level synthesis software from Xilinx that translate C or C++ code into Verilog/VHDL and packages the resulting kernel into an IP core. Vivado IPI (IP integrator) is instead an EDA tool to connect, synthesize and place these IP blocks for a specified FPGA model (in our case the Xilinx Zynq-7020 included in Zedboard). It can also acts as a standard FPGA tool by directly writing HDL code for the blocks and the connections. Finally, Xilinx SDK was used to run the management software on the on-board ARM processor, used to manage the pictures and to send and receive them from the programmable logic.
In short, the FPGA implementation outperformed (by 6X) the sofwtare implementation (running on a 2012 Intel core-i7), and was also more energy efficient. We compared the work also the other state of art implementations and ours was faster! This allowed us to arrive to the Xilinx Open Hardware competition finals, and to publish a paper in the EMBC 2017 conference (a high-ranked biomedical conference) in South Korea.
Media and Contacts
Xilinx Open Hardware 2016 Finalists - link
Source Code, designs etc… - Bitbucket
Some videos explaining the project - YouTube playlist
Paper published at EMBC 2017 - pdf, poster, ieee
Marco Bacis - [email protected]
Lara Cavinato - [email protected]
Irene Fidone - [email protected]
Marco D. Santambrogio - [email protected]
And here are a few slides showing the project (presented at the Stanford OTL office in May 2018):